Project funded by National Institutes of Health combines mathematics and biology to identify cellular forces to target cancer
Two researchers at Worcester Polytechnic Institute (WPI) have been awarded $917,999 by the National Institutes of Health to develop computational models for the study of a critical piece of cellular machinery that often goes awry in cancer.
The three-year study will use mathematical techniques and biological findings to assess how cellular forces influence the geometry of the mitotic spindle, a part of the cell’s machinery that is responsible for separating genetic material during cell division.
“There is too much going on during cell division to tease out and examine all the possible forces at work through laboratory experiments,” said Sarah Olson, WPI associate professor of mathematical sciences and the principal investigator (PI) of the project. “But by combining experiments with modeling, you can explore factors that lead to defective spindle structure in cells.” 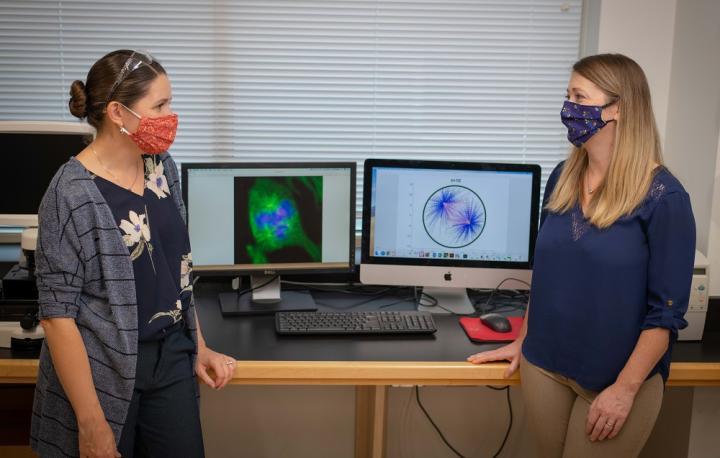 {module INSIDE STORY}
{module INSIDE STORY}
The study will build on previous work done by Olson and co-PI Amity Manning, assistant professor of biology and biotechnology, to build computational models to illuminate the forces in human epithelial cells during division. Both Olson and Manning are affiliated with WPI’s Bioinformatics and Computational Biology program.
Computational models use math and supercomputer simulations to adjust numerous variables in a complex system and observe outcomes. Cell division is a complex process in which a parent cell makes a copy of its chromosomes, which contain genetic instructions, and then splits into two new daughter cells.
A healthy cell undergoing division contains two centrosomes that anchor opposite ends of a molecular spindle. As the cell divides, one copy of all chromosomes is pulled toward each anchor point or spindle pole. Both cells that result from the division inherit an identical complement of chromosomes.
Cancer cells, however, often contain more than two centrosomes. The extra centrosomes must cluster together to form a functional spindle with two spindle poles so that a cancer cell can divide into two new cancer cells. If the extra centrosomes do not cluster together, a cancer cell with extra centrosomes divides into more than two new cells, each of which inherits too little genetic material to survive. This suggests that interventions that limit centrosome clustering could promote the death of cancer cells.
Researchers can watch cell division under microscopes, manipulate genes and proteins involved in the process, and monitor the consequences when defects occur, but there are limits to how much can be accomplished in laboratory experiments, Manning said.
“There is a lot of redundancy and overlapping functions in cell division,” Manning said. “We want to understand how centrosome clustering is regulated and how that influences basic cell biology. With modeling, we can simplify complex functions and test scenarios to better understand what’s happening.”
During the project, the researchers will develop new computational models and elucidate the relationship between initial centrosome positions in a cell and cell division. They also will identify how forces, such as the motor protein dynein, impact the movement of centrosomes in a dividing cell with extra centrosomes. Laboratory experiments will inform new computational models, Olson and Manning said, and the models will spur additional laboratory experiments.
“This is truly a collaborative project with a balance of math and biology,” Manning said. “We can brainstorm and think about modeling questions, biological questions, and how we can apply our expertise to this problem.”


 How to resolve AdBlock issue?
How to resolve AdBlock issue?