ENGINEERING
Finnish university develops ML approach that facilitates molecular conformer search in complex molecules
At the Computational Electronic Structure Theory Group (CEST) at Aalto University in Finland, researchers have developed a new machine learning approach based on a low-energy latent space (LOLS) and density functional theory (DFT) to search for molecular conformers. 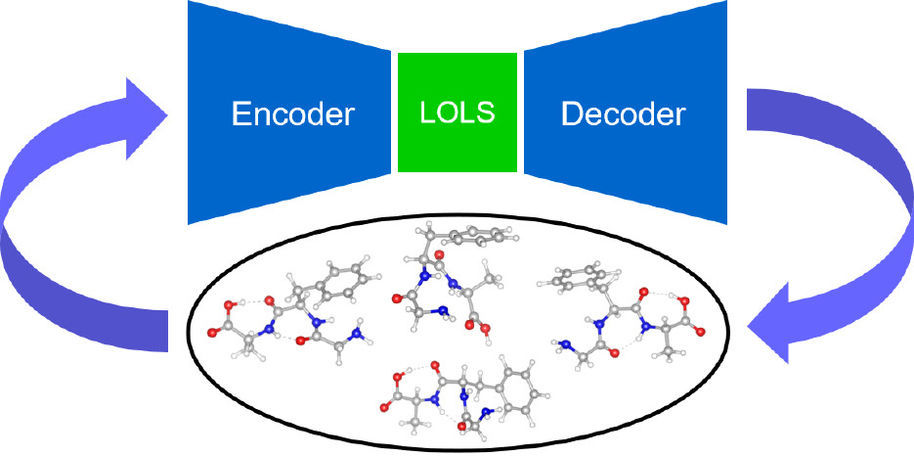
Molecular conformer search is a topic of great importance in computational chemistry, drug design, and material science. The challenge is to identify low-energy conformers in the first place. This difficulty arises from the high complexity of search spaces, as well as the computational cost associated with accurate quantum chemical methods. In the past, conformer search would take up considerable time and computational resources.
To address this challenge, visiting doctoral student Xiaomi Guo, together with other CEST researchers Lincan Fang, Prof. Patrick Rinke, Dr. Xi Chen, and Prof. Milica Todorovic (University of Turku) explored the possibility of performing the molecular conformer search in a low-dimensional latent space. This method uses a generative model variational auto-encoder (VAE) and biases the VAE towards low-energy molecular configurations to generate more informative data. In this way, the model can effectively learn the low-energy potential surface and hence identify the related molecular conformers. The CEST teams call their new method low-energy latent space (LOLS) conformer search.
In a recent publication, the authors tested this new LOLS procedure on amino acids and peptides with 5–9 searching dimensions. The new results agree well with previous studies. The team found that for small molecules such as cysteine, it is more efficient to sample data in real space; however, LOLS turns out to be more suitable for larger molecules such as peptides. The authors now plan to extend their structure search methods to more complex materials beyond molecules.

